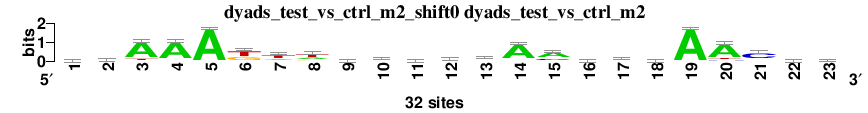
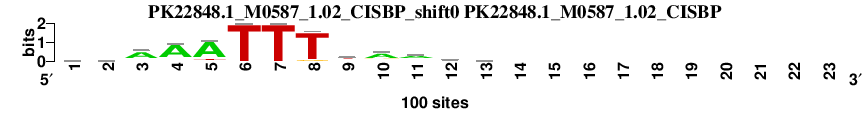
| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SSD |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSSD |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
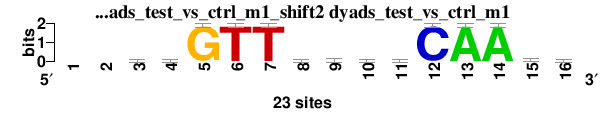
| dyads_test_vs_ctrl_m1_shift2 (dyads_test_vs_ctrl_m1) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; dyads_test_vs_ctrl_m1; m=0 (reference); ncol1=14; shift=2; ncol=16; --bwGTTbtwtCAAam
; Alignment reference
a 0 0 3 8 0 0 0 5 4 9 5 0 23 23 10 9
c 0 0 6 5 0 0 0 6 5 3 4 23 0 0 3 6
g 0 0 7 3 23 0 0 6 4 5 5 0 0 0 5 3
t 0 0 7 7 0 23 23 6 10 6 9 0 0 0 5 5
|
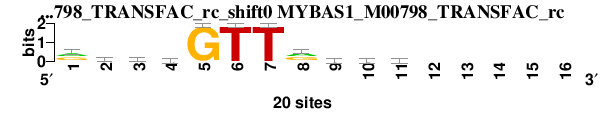
| MYBAS1_M00798_TRANSFAC_rc_shift0 (MYBAS1_M00798_TRANSFAC_rc) |
 |
0.884 |
0.497 |
0.018 |
-0.070 |
0.940 |
0.587 |
0.967 |
2 |
1 |
6 |
5 |
1 |
2 |
1 |
2.571 |
1 |
; dyads_test_vs_ctrl_m1 versus MYBAS1_M00798_TRANSFAC_rc; m=1/5; ncol2=11; w=9; offset=-2; strand=R; shift=0; score= 2.5714; rgcmGTTrbsg-----
; cor=0.884; Ncor=0.497; logoDP=0.018; NIcor=-0.070; NsEucl=0.940; SSD=0.587; NSW=0.967; rcor=2; rNcor=1; rlogoDP=6; rNIcor=5; rNsEucl=1; rSSD=2; rNSW=1; rank_mean=2.571; match_rank=1
a 8 4 4 6 0 0 0 9 2 2 2 0 0 0 0 0
c 0 3 9 6 0 0 0 2 5 5 4 0 0 0 0 0
g 8 9 4 4 20 0 0 9 6 7 7 0 0 0 0 0
t 2 3 2 3 0 20 20 0 5 4 4 0 0 0 0 0
|
| MYB2_MYB2_Athamap_rc_shift1 (MYB2_MYB2_Athamap_rc) |
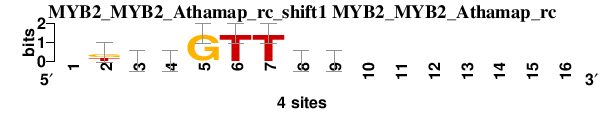 |
0.899 |
0.419 |
0.285 |
-0.026 |
0.926 |
0.531 |
0.962 |
1 |
5 |
3 |
3 |
3 |
1 |
2 |
2.571 |
2 |
; dyads_test_vs_ctrl_m1 versus MYB2_MYB2_Athamap_rc; m=2/5; ncol2=8; w=7; offset=-1; strand=R; shift=1; score= 2.5714; -kbvGTTdb-------
; cor=0.899; Ncor=0.419; logoDP=0.285; NIcor=-0.026; NsEucl=0.926; SSD=0.531; NSW=0.962; rcor=1; rNcor=5; rlogoDP=3; rNIcor=3; rNsEucl=3; rSSD=1; rNSW=2; rank_mean=2.571; match_rank=2
a 0 0 0 2 0 0 0 2 0 0 0 0 0 0 0 0
c 0 0 2 1 0 0 0 0 1 0 0 0 0 0 0 0
g 0 2 1 1 4 0 0 1 2 0 0 0 0 0 0 0
t 0 2 1 0 0 4 4 1 1 0 0 0 0 0 0 0
|
| ZFP5_M0369_1.02_CISBP_rc_shift3 (ZFP5_M0369_1.02_CISBP_rc) |
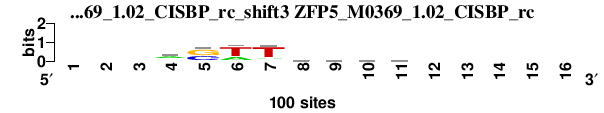 |
0.817 |
0.467 |
0.031 |
-0.049 |
0.917 |
0.880 |
0.945 |
3 |
3 |
5 |
4 |
4 |
3 |
3 |
3.571 |
4 |
; dyads_test_vs_ctrl_m1 versus ZFP5_M0369_1.02_CISBP_rc; m=3/5; ncol2=8; w=8; offset=1; strand=R; shift=3; score= 3.5714; ---aswThvmw-----
; cor=0.817; Ncor=0.467; logoDP=0.031; NIcor=-0.049; NsEucl=0.917; SSD=0.880; NSW=0.945; rcor=3; rNcor=3; rlogoDP=5; rNIcor=4; rNsEucl=4; rSSD=3; rNSW=3; rank_mean=3.571; match_rank=4
a 0 0 0 53 1 28 11 25 27 28 26 0 0 0 0 0
c 0 0 0 6 41 6 9 31 25 27 24 0 0 0 0 0
g 0 0 0 22 53 0 5 18 25 24 24 0 0 0 0 0
t 0 0 0 19 5 66 75 26 23 21 26 0 0 0 0 0
|
| ZFP8_M0371_1.02_CISBP_shift3 (ZFP8_M0371_1.02_CISBP) |
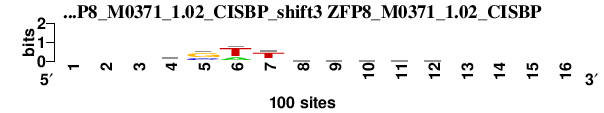 |
0.764 |
0.491 |
1.129 |
0.510 |
0.902 |
1.556 |
0.914 |
5 |
2 |
2 |
1 |
6 |
5 |
6 |
3.857 |
5 |
; dyads_test_vs_ctrl_m1 versus ZFP8_M0371_1.02_CISBP; m=4/5; ncol2=9; w=9; offset=1; strand=D; shift=3; score= 3.8571; ---wswthvmdy----
; cor=0.764; Ncor=0.491; logoDP=1.129; NIcor=0.510; NsEucl=0.902; SSD=1.556; NSW=0.914; rcor=5; rNcor=2; rlogoDP=2; rNIcor=1; rNsEucl=6; rSSD=5; rNSW=6; rank_mean=3.857; match_rank=5
a 0 0 0 46 4 27 14 26 27 27 25 24 0 0 0 0
c 0 0 0 14 32 8 10 29 25 26 23 25 0 0 0 0
g 0 0 0 14 56 0 10 19 25 24 26 22 0 0 0 0
t 0 0 0 26 8 65 66 26 23 23 26 29 0 0 0 0
|
| Gam1_MA0034.1_JASPAR_rc_shift0 (Gam1_MA0034.1_JASPAR_rc) |
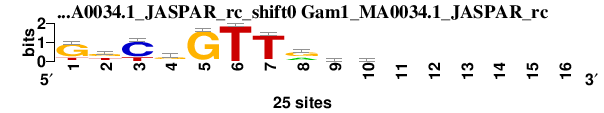 |
0.808 |
0.404 |
0.073 |
-0.084 |
0.911 |
1.011 |
0.937 |
4 |
6 |
4 |
6 |
5 |
4 |
5 |
4.857 |
6 |
; dyads_test_vs_ctrl_m1 versus Gam1_MA0034.1_JASPAR_rc; m=5/5; ncol2=10; w=8; offset=-2; strand=R; shift=0; score= 4.8571; GkCgGTTryc------
; cor=0.808; Ncor=0.404; logoDP=0.073; NIcor=-0.084; NsEucl=0.911; SSD=1.011; NSW=0.937; rcor=4; rNcor=6; rlogoDP=4; rNIcor=6; rNsEucl=5; rSSD=4; rNSW=5; rank_mean=4.857; match_rank=6
a 0 4 0 2 0 0 0 9 3 4 0 0 0 0 0 0
c 1 0 19 6 0 0 1 0 7 11 0 0 0 0 0 0
g 19 11 0 14 24 0 1 13 5 6 0 0 0 0 0 0
t 5 10 6 3 1 25 23 3 10 4 0 0 0 0 0 0
|