RSAT - matrix-quality result
Analysis: matrix-quality result (21/03/2018 17:50)
matrix-quality result: LexA and CTCF matrices from RegulonDB 2015
Command: matrix-quality -v 0 -html_title ' LexA and CTCF matrices from RegulonDB 2015 ' -ms $RSAT/public_html/tmp/apache/2018/03/21/matrix-quality_2018-03-21.175002_zdBRpm/input_matrix -matrix_format transfac -pseudo 1 -seq LexA_peaks $RSAT/public_html/tmp/apache/2018/03/21/matrix-quality_2018-03-21.175002_zdBRpm/sequence1.fasta -seq_format fasta -plot LexA_peaks nwd -seq CRP_binding_sites $RSAT/public_html/tmp/apache/2018/03/21/matrix-quality_2018-03-21.175002_zdBRpm/sequence2.fasta -plot CRP_binding_sites nwd -perm LexA_peaks 1 -perm CRP_binding_sites 1 -bgfile $RSAT/public_html/data/genomes/Escherichia_coli_GCF_000005845.2_ASM584v2/oligo-frequencies/2nt_upstream-noorf_Escherichia_coli_GCF_000005845.2_ASM584v2-ovlp-1str.freq.gz -bg_format oligo-analysis -bg_pseudo 0.01 -archive -o $RSAT/public_html/tmp/apache/2018/03/21/matrix-quality_2018-03-21.175002_zdBRpm/matrix-quality_2018-03-21.175002
| Matrix name | Logo | HTML report |
|---|
| LexA.RegulonDB2012 | 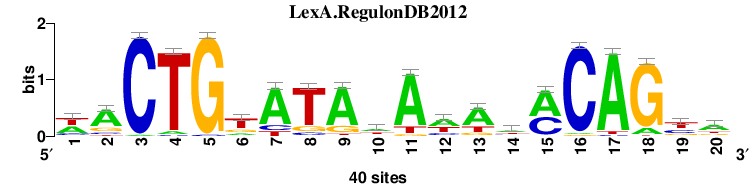
| report |
| CRP.RegulonDB2012 | 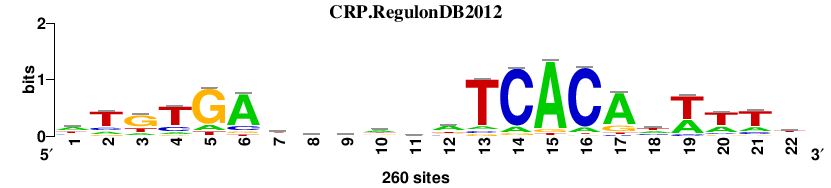
| report |
NWD curves
NWD comparison heatmaps
| 

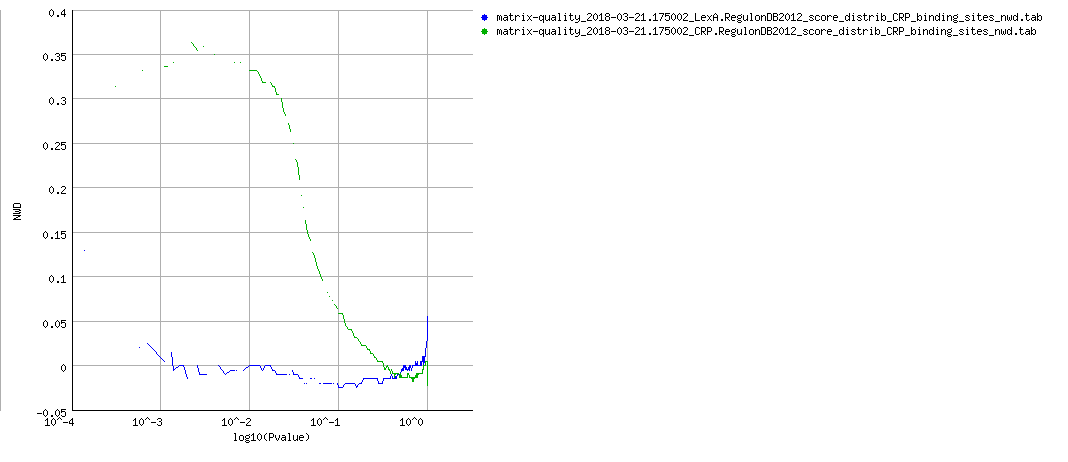
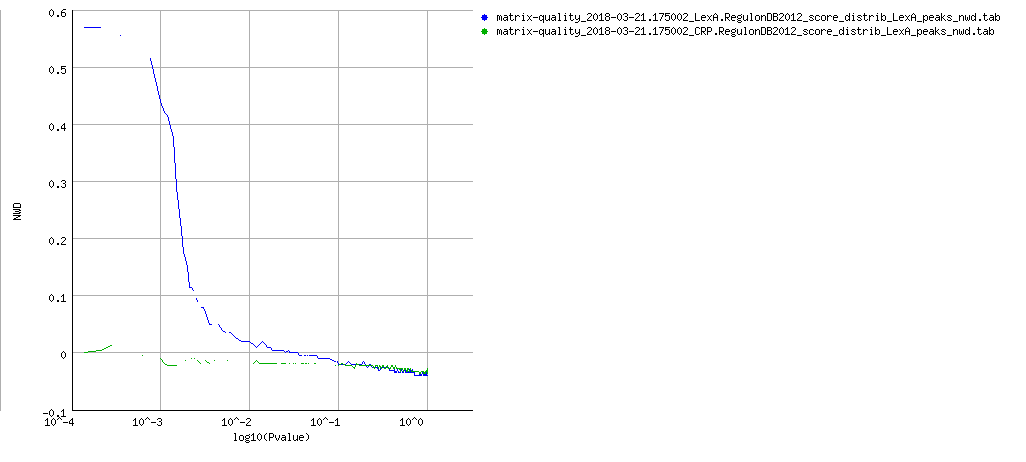
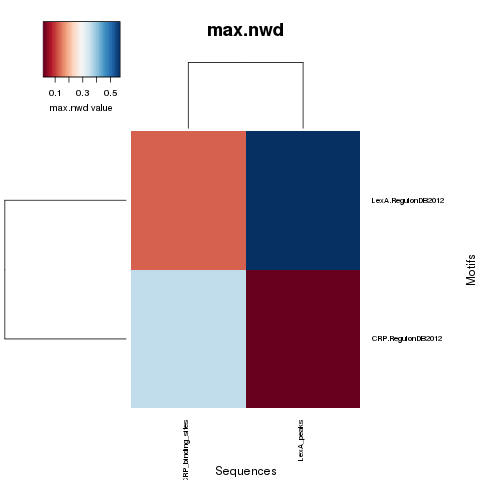 PDF Table
PDF Table 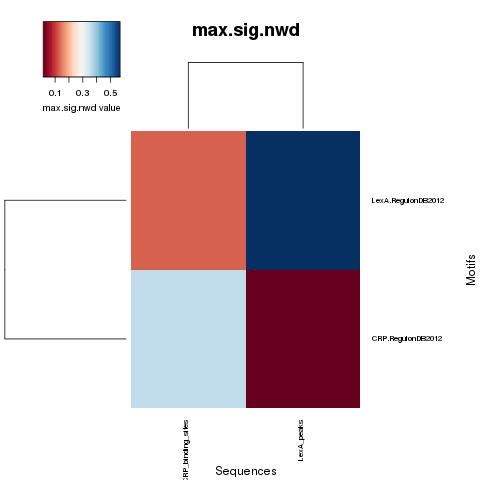 PDF Table
PDF Table 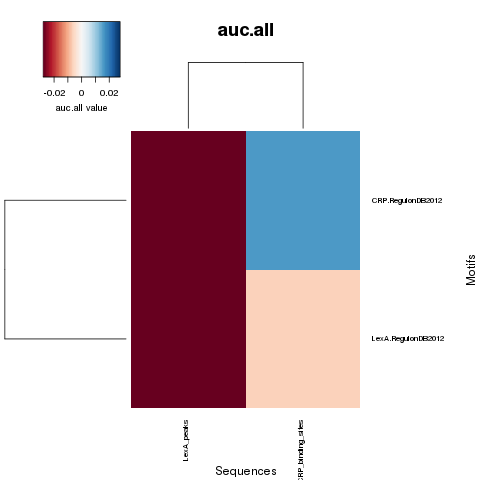 PDF Table
PDF Table 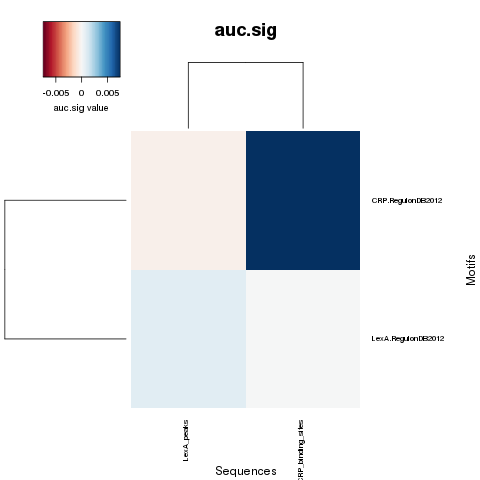 PDF Table
PDF Table