One-to-n alignments
Command: compare-matrices -v 1 -format1 transfac -file1 $RSAT/public_html/tmp/www-data/2025/03/26/compare-matrices_2025-03-26.111727_8jZM1g/compare-matrices_query_matrices.transfac -file2 $RSAT/public_html/motif_databases/JASPAR/Jaspar_2024/JASPAR2024_CORE_fungi_non-redundant_pfms_transfac.txt -format2 tf -strand DR -lth cor 0.7 -lth Ncor 0.4 -uth match_rank 50 -return cor,Ncor,logoDP,NIcor,NsEucl,SW,NSW,offset,match_rank,matrix_id,matrix_name,width,strand,offset,consensus,alignments_1ton -o $RSAT/public_html/tmp/www-data/2025/03/26/compare-matrices_2025-03-26.111727_8jZM1g/compare-matrices.tab
One-to-n matrix alignment; reference matrix: positions_7nt_m3_shift0 ; 2 matrices ; sort_field=rank_mean
| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SW |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSW |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
| positions_7nt_m3_shift0 (positions_7nt_m3) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; positions_7nt_m3; m=0 (reference); ncol1=11; shift=0; ncol=11; AAAAAAAAAAA
; Alignment reference
a 457 495 663 661 662 662 663 662 663 498 460
c 51 51 0 0 0 0 0 0 0 55 63
g 79 69 0 0 0 0 0 0 0 55 73
t 76 48 0 2 1 1 0 1 0 55 67
|
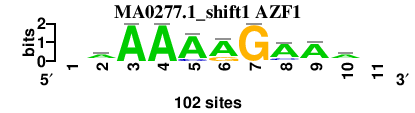
| MA0277.1_shift1 (AZF1) |
 |
0.782 |
0.640 |
7.566 |
0.637 |
0.880 |
15.677 |
0.871 |
4 |
1 |
1 |
1 |
1 |
1 |
4 |
1.857 |
1 |
; positions_7nt_m3 versus MA0277.1 (AZF1); m=1/1; ncol2=9; w=0; offset=1; strand=D; shift=1; score= 1.8571; -aAAARGAAa-
; cor=0.782; Ncor=0.640; logoDP=7.566; NIcor=0.637; NsEucl=0.880; SW=15.677; NSW=0.871; rcor=4; rNcor=1; rlogoDP=1; rNIcor=1; rNsEucl=1; rSW=1; rNSW=4; rank_mean=1.857; match_rank=1
a 0 63.0 100.0 100.0 88.0 75.0 0.0 78.0 81.0 63.0 0
c 0 13.0 0.0 0.0 13.0 0.0 0.0 16.0 6.0 13.0 0
g 0 13.0 0.0 0.0 0.0 25.0 100.0 3.0 6.0 13.0 0
t 0 13.0 0.0 0.0 0.0 0.0 0.0 3.0 6.0 13.0 0
|
One-to-n matrix alignment; reference matrix: oligos_7nt_mkv5_m2_shift0 ; 4 matrices ; sort_field=rank_mean
| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SW |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSW |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
| oligos_7nt_mkv5_m2_shift0 (oligos_7nt_mkv5_m2) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; oligos_7nt_mkv5_m2; m=0 (reference); ncol1=12; shift=0; ncol=12; wtATGCAAATgw
; Alignment reference
a 251 131 562 0 6 2 417 423 563 3 134 181
c 38 102 1 3 0 468 3 10 1 13 74 115
g 69 72 2 0 554 2 7 101 1 5 241 109
t 207 260 0 562 5 93 138 31 0 544 116 160
|
| MA0355.2_shift1 (PHD1) |
 |
0.822 |
0.411 |
4.641 |
0.404 |
0.879 |
10.940 |
0.912 |
2 |
6 |
4 |
2 |
2 |
4 |
1 |
3.000 |
2 |
; oligos_7nt_mkv5_m2 versus MA0355.2 (PHD1); m=1/3; ncol2=6; w=0; offset=1; strand=D; shift=1; score= 3; -smTGCA-----
; cor=0.822; Ncor=0.411; logoDP=4.641; NIcor=0.404; NsEucl=0.879; SW=10.940; NSW=0.912; rcor=2; rNcor=6; rlogoDP=4; rNIcor=2; rNsEucl=2; rSW=4; rNSW=1; rank_mean=3.000; match_rank=2
a 0 9.0 37.0 0.0 0.0 0.0 91.0 0 0 0 0 0
c 0 43.0 44.0 10.0 0.0 99.0 1.0 0 0 0 0 0
g 0 39.0 15.0 0.0 99.0 0.0 7.0 0 0 0 0 0
t 0 8.0 3.0 89.0 0.0 1.0 1.0 0 0 0 0 0
|
| MA0274.1_rc_shift2 (ARR1_rc) |
 |
0.755 |
0.503 |
4.068 |
0.319 |
0.873 |
13.929 |
0.871 |
5 |
2 |
5 |
5 |
3 |
2 |
5 |
3.857 |
4 |
; oligos_7nt_mkv5_m2 versus MA0274.1_rc (ARR1_rc); m=2/3; ncol2=8; w=0; offset=2; strand=R; shift=2; score= 3.8571; --ATTYArrT--
; cor=0.755; Ncor=0.503; logoDP=4.068; NIcor=0.319; NsEucl=0.873; SW=13.929; NSW=0.871; rcor=5; rNcor=2; rlogoDP=5; rNIcor=5; rNsEucl=3; rSW=2; rNSW=5; rank_mean=3.857; match_rank=4
a 0 0 129.0 3.0 0.0 0.0 161.0 87.0 73.0 0.0 0 0
c 0 0 24.0 29.0 12.0 161.0 0.0 0.0 29.0 0.0 0 0
g 0 0 0.0 0.0 41.0 0.0 0.0 70.0 75.0 0.0 0 0
t 0 0 11.0 137.0 129.0 73.0 0.0 30.0 24.0 160.0 0 0
|
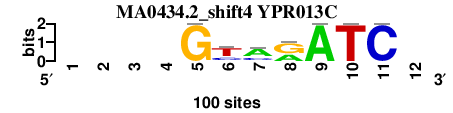
| MA0434.2_shift4 (YPR013C) |
 |
0.719 |
0.419 |
5.745 |
0.388 |
0.857 |
11.999 |
0.857 |
6 |
4 |
2 |
3 |
6 |
3 |
6 |
4.286 |
5 |
; oligos_7nt_mkv5_m2 versus MA0434.2 (YPR013C); m=3/3; ncol2=7; w=0; offset=4; strand=D; shift=4; score= 4.2857; ----GymrATC-
; cor=0.719; Ncor=0.419; logoDP=5.745; NIcor=0.388; NsEucl=0.857; SW=11.999; NSW=0.857; rcor=6; rNcor=4; rlogoDP=2; rNIcor=3; rNsEucl=6; rSW=3; rNSW=6; rank_mean=4.286; match_rank=5
a 0 0 0 0 0.0 9.0 63.0 40.0 100.0 0.0 0.0 0
c 0 0 0 0 0.0 26.0 26.0 0.0 0.0 0.0 100.0 0
g 0 0 0 0 100.0 0.0 0.0 60.0 0.0 0.0 0.0 0
t 0 0 0 0 0.0 65.0 11.0 0.0 0.0 100.0 0.0 0
|
One-to-n matrix alignment; reference matrix: oligos_7nt_mkv5_m3_shift0 ; 3 matrices ; sort_field=rank_mean
| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SW |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSW |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
| oligos_7nt_mkv5_m3_shift0 (oligos_7nt_mkv5_m3) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; oligos_7nt_mkv5_m3; m=0 (reference); ncol1=11; shift=0; ncol=11; wtATGyTAAtk
; Alignment reference
a 95 61 246 0 0 1 19 245 246 53 60
c 25 46 0 2 0 166 0 0 0 35 32
g 33 47 0 0 245 0 1 0 0 31 75
t 93 92 0 244 1 79 226 1 0 127 79
|
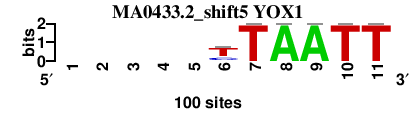
| MA0433.2_shift5 (YOX1) |
 |
0.829 |
0.414 |
5.484 |
0.380 |
0.868 |
10.738 |
0.895 |
1 |
5 |
3 |
4 |
4 |
5 |
2 |
3.429 |
3 |
; oligos_7nt_mkv5_m3 versus MA0433.2 (YOX1); m=1/2; ncol2=7; w=0; offset=5; strand=D; shift=5; score= 3.4286; -----yTAATT
; cor=0.829; Ncor=0.414; logoDP=5.484; NIcor=0.380; NsEucl=0.868; SW=10.738; NSW=0.895; rcor=1; rNcor=5; rlogoDP=3; rNIcor=4; rNsEucl=4; rSW=5; rNSW=2; rank_mean=3.429; match_rank=3
a 0 0 0 0 0 1.0 0.0 100.0 100.0 0.0 0.0
c 0 0 0 0 0 25.0 0.0 0.0 0.0 0.0 0.0
g 0 0 0 0 0 7.0 0.0 0.0 0.0 0.0 0.0
t 0 0 0 0 0 66.0 100.0 0.0 0.0 100.0 100.0
|
| MA0417.1_rc_shift2 (YAP5_rc) |
 |
0.793 |
0.433 |
2.375 |
0.072 |
0.858 |
10.543 |
0.879 |
3 |
3 |
6 |
6 |
5 |
6 |
3 |
4.571 |
6 |
; oligos_7nt_mkv5_m3 versus MA0417.1_rc (YAP5_rc); m=2/2; ncol2=6; w=0; offset=2; strand=R; shift=2; score= 4.5714; --ATGyyT---
; cor=0.793; Ncor=0.433; logoDP=2.375; NIcor=0.072; NsEucl=0.858; SW=10.543; NSW=0.879; rcor=3; rNcor=3; rlogoDP=6; rNIcor=6; rNsEucl=5; rSW=6; rNSW=3; rank_mean=4.571; match_rank=6
a 0 0 137.0 0.0 17.0 0.0 0.0 34.0 0 0 0
c 0 0 0.0 0.0 0.0 136.0 67.0 0.0 0 0 0
g 0 0 0.0 0.0 264.0 0.0 0.0 0.0 0 0 0
t 0 0 19.0 161.0 0.0 86.0 124.0 122.0 0 0 0
|








