One-to-n alignments
Command: compare-matrices -v 1 -format1 transfac -file1 $RSAT/public_html/tmp/www-data/2025/09/15/compare-matrices_2025-09-15.163721_GCEzmB/compare-matrices_query_matrices.transfac -file2 $RSAT/public_html/motif_databases/ArabidopsisPBM/ArabidopsisPBM_2015-11-06.tf -format2 tf -strand DR -lth cor 0.7 -lth Ncor 0.4 -uth match_rank 50 -return cor,Ncor,logoDP,NsEucl,NSW,match_rank,matrix_id,matrix_name,width,strand,offset,consensus,alignments_1ton -o $RSAT/public_html/tmp/www-data/2025/09/15/compare-matrices_2025-09-15.163721_GCEzmB/compare-matrices.tab
One-to-n matrix alignment; reference matrix: tcp4_c1_m11_assembly_11_shift0 ; 2 matrices ; sort_field=rank_mean
| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NsEucl |
NSW |
rcor |
rNcor |
rlogoDP |
rNsEucl |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
| tcp4_c1_m11_assembly_11_shift0 (assembly_11) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
; tcp4_c1_m11_assembly_11 (assembly_11); m=0 (reference); ncol1=12; shift=0; ncol=12; wwTGAGCTGAww
; Alignment reference
a 116 146 20 15 409 22 38 18 30 371 147 159
c 62 61 12 6 2 12 344 4 18 17 64 59
g 79 62 15 367 7 357 11 3 350 11 77 89
t 165 153 375 34 4 31 29 397 24 23 134 115
|
| 7207_At4g35610_ArabidopsisPBM_20140210__shift1 (7207_At4g35610_ArabidopsisPBM_20140210_) |
 |
0.795 |
0.662 |
2.898 |
0.911 |
0.920 |
1 |
1 |
5 |
1 |
1 |
1.800 |
1 |
; tcp4_c1_m11_assembly_11 versus 7207_At4g35610_ArabidopsisPBM_20140210_; m=1/1; ncol2=10; w=0; offset=1; strand=D; shift=1; score= 1.8; -rkCAGCTGmm-
; cor=0.795; Ncor=0.662; logoDP=2.898; NsEucl=0.911; NSW=0.920; rcor=1; rNcor=1; rlogoDP=5; rNsEucl=1; rNSW=1; rank_mean=1.800; match_rank=1
a 0 31 13 8 83 3 8 10 11 55 38 0
c 0 14 6 68 5 2 87 2 13 26 38 0
g 0 36 26 13 2 87 2 5 68 6 15 0
t 0 19 55 11 10 8 3 83 8 13 9 0
|
One-to-n matrix alignment; reference matrix: tcp4_c1_m5_assembly_5_shift1 ; 2 matrices ; sort_field=rank_mean
| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NsEucl |
NSW |
rcor |
rNcor |
rlogoDP |
rNsEucl |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
| tcp4_c1_m5_assembly_5_shift1 (assembly_5) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
; tcp4_c1_m5_assembly_5 (assembly_5); m=0 (reference); ncol1=11; shift=1; ncol=12; -hdCCGGCGAww
; Alignment reference
a 0 127 122 30 13 16 21 38 21 371 139 127
c 0 106 50 336 376 3 10 337 19 11 83 63
g 0 62 112 24 8 379 358 8 352 15 71 100
t 0 117 128 22 15 14 23 29 20 15 119 122
|
| 7203_ATERF1_2_ArabidopsisPBM_20140210__shift0 (7203_ATERF1_2_ArabidopsisPBM_20140210_) |
 |
0.737 |
0.553 |
3.906 |
0.888 |
0.887 |
3 |
3 |
4 |
2 |
2 |
2.800 |
2 |
; tcp4_c1_m5_assembly_5 versus 7203_ATERF1_2_ArabidopsisPBM_20140210_; m=1/1; ncol2=10; w=-1; offset=-1; strand=D; shift=0; score= 2.8; ytGCCGGCar--
; cor=0.737; Ncor=0.553; logoDP=3.906; NsEucl=0.888; NSW=0.887; rcor=3; rNcor=3; rlogoDP=4; rNsEucl=2; rNSW=2; rank_mean=2.800; match_rank=2
a 14 14 4 3 1 4 1 1 60 35 0 0
c 32 14 0 90 95 1 6 95 11 20 0 0
g 24 12 95 6 0 94 90 0 15 29 0 0
t 30 60 1 1 4 1 3 4 14 16 0 0
|
One-to-n matrix alignment; reference matrix: tcp4_c1_m16_assembly_16_shift0 ; 2 matrices ; sort_field=rank_mean
| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NsEucl |
NSW |
rcor |
rNcor |
rlogoDP |
rNsEucl |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
| tcp4_c1_m16_assembly_16_shift0 (assembly_16) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
; tcp4_c1_m16_assembly_16 (assembly_16); m=0 (reference); ncol1=10; shift=0; ncol=10; wwCAAAAGww
; Alignment reference
a 410 391 0 1043 1043 1043 1043 0 413 397
c 170 191 1043 0 0 0 0 0 149 155
g 190 147 0 0 0 0 0 1043 176 167
t 273 314 0 0 0 0 0 0 305 324
|
| 7217_DOF5.7_ArabidopsisPBM_20140210__shift2 (7217_DOF5.7_ArabidopsisPBM_20140210_) |
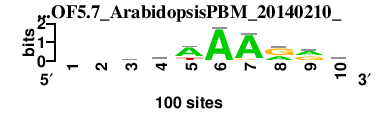 |
0.738 |
0.492 |
3.989 |
0.872 |
0.869 |
2 |
4 |
2 |
4 |
3 |
3.000 |
3 |
; tcp4_c1_m16_assembly_16 versus 7217_DOF5.7_ArabidopsisPBM_20140210_; m=1/1; ncol2=10; w=0; offset=2; strand=D; shift=2; score= 3; --raAAArrk
; cor=0.738; Ncor=0.492; logoDP=3.989; NsEucl=0.872; NSW=0.869; rcor=2; rNcor=4; rlogoDP=2; rNsEucl=4; rNSW=3; rank_mean=3.000; match_rank=3
a 0 0 39 45 68 97 90 36 48 10
c 0 0 21 9 1 0 0 4 6 19
g 0 0 27 22 7 1 8 59 44 46
t 0 0 13 24 24 2 2 1 2 25
|
One-to-n matrix alignment; reference matrix: tcp4_c1_m14_assembly_14_shift0 ; 3 matrices ; sort_field=rank_mean
| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NsEucl |
NSW |
rcor |
rNcor |
rlogoDP |
rNsEucl |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
| tcp4_c1_m14_assembly_14_shift0 (assembly_14) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
; tcp4_c1_m14_assembly_14 (assembly_14); m=0 (reference); ncol1=10; shift=0; ncol=10; waAGAAAGww
; Alignment reference
a 381 440 951 0 951 951 951 0 461 373
c 146 100 0 0 0 0 0 0 123 114
g 178 231 0 951 0 0 0 951 123 210
t 246 180 0 0 0 0 0 0 244 254
|
| 7211_DAG2_ArabidopsisPBM_20140210__shift1 (7211_DAG2_ArabidopsisPBM_20140210_) |
 |
0.703 |
0.575 |
6.857 |
0.875 |
0.860 |
5 |
2 |
1 |
3 |
5 |
3.200 |
4 |
; tcp4_c1_m14_assembly_14 versus 7211_DAG2_ArabidopsisPBM_20140210_; m=1/2; ncol2=10; w=0; offset=1; strand=D; shift=1; score= 3.2; -sAwAARGTg
; cor=0.703; Ncor=0.575; logoDP=6.857; NsEucl=0.875; NSW=0.860; rcor=5; rNcor=2; rlogoDP=1; rNsEucl=3; rNSW=5; rank_mean=3.200; match_rank=4
a 0 16 75 60 99 98 75 0 1 24
c 0 41 3 0 0 0 0 0 20 4
g 0 31 18 1 0 1 25 98 6 56
t 0 12 4 39 1 1 0 2 73 16
|
| 7217_DOF5.7_ArabidopsisPBM_20140210__shift2 (7217_DOF5.7_ArabidopsisPBM_20140210_) |
 |
0.730 |
0.487 |
3.975 |
0.870 |
0.864 |
4 |
5 |
3 |
5 |
4 |
4.200 |
5 |
; tcp4_c1_m14_assembly_14 versus 7217_DOF5.7_ArabidopsisPBM_20140210_; m=2/2; ncol2=10; w=0; offset=2; strand=D; shift=2; score= 4.2; --raAAArrk
; cor=0.730; Ncor=0.487; logoDP=3.975; NsEucl=0.870; NSW=0.864; rcor=4; rNcor=5; rlogoDP=3; rNsEucl=5; rNSW=4; rank_mean=4.200; match_rank=5
a 0 0 39 45 68 97 90 36 48 10
c 0 0 21 9 1 0 0 4 6 19
g 0 0 27 22 7 1 8 59 44 46
t 0 0 13 24 24 2 2 1 2 25
|







