| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NsEucl |
NSW |
rcor |
rNcor |
rlogoDP |
rNsEucl |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
| cluster_5_shift0 (cluster_5) |
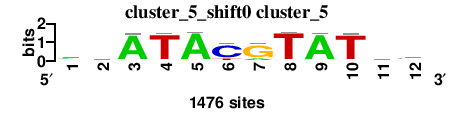 |
|
|
|
|
|
|
|
|
|
|
|
|
; cluster_5; m=0 (reference); ncol1=12; shift=0; ncol=12; wwATACGTATww
; Alignment reference
a 682 442 1350 64 1375 82 247 46 1353 53 560 404
c 179 267 32 42 30 1117 30 25 17 41 207 211
g 211 207 41 17 25 30 1117 30 42 32 267 179
t 404 560 53 1353 46 247 82 1375 64 1350 442 682
|
| bZIP65_EEAD0071_EEADannot_shift3 (bZIP65:EEAD0071:EEADannot) |
 |
0.988 |
0.494 |
6.670 |
0.946 |
0.982 |
1 |
4 |
1 |
1 |
1 |
1.600 |
1 |
; cluster_5 versus bZIP65_EEAD0071_EEADannot (bZIP65:EEAD0071:EEADannot); m=1/6; ncol2=6; w=0; offset=3; strand=D; shift=3; score= 1.6; ---TACGTA---
; cor=0.988; Ncor=0.494; logoDP=6.670; NsEucl=0.946; NSW=0.982; rcor=1; rNcor=4; rlogoDP=1; rNsEucl=1; rNSW=1; rank_mean=1.600; match_rank=1
a 0 0 0 0 1 0 0 0 1 0 0 0
c 0 0 0 0 0 1 0 0 0 0 0 0
g 0 0 0 0 0 0 1 0 0 0 0 0
t 0 0 0 1 0 0 0 1 0 0 0 0
|
| ANAC55_ANAC55_2_ArabidopsisPBM_shift1 (ANAC55:ANAC55_2:ArabidopsisPBM) |
 |
0.740 |
0.617 |
3.436 |
0.898 |
0.896 |
4 |
1 |
4 |
2 |
4 |
3.000 |
2 |
; cluster_5 versus ANAC55_ANAC55_2_ArabidopsisPBM (ANAC55:ANAC55_2:ArabidopsisPBM); m=2/6; ncol2=10; w=0; offset=1; strand=D; shift=1; score= 3; -rwTACGTAwy-
; cor=0.740; Ncor=0.617; logoDP=3.436; NsEucl=0.898; NSW=0.896; rcor=4; rNcor=1; rlogoDP=4; rNsEucl=2; rNSW=4; rank_mean=3.000; match_rank=2
a 0 39 27 3 89 5 5 2 70 64 6 0
c 0 11 3 22 5 88 2 4 4 6 32 0
g 0 30 6 4 4 2 88 5 23 3 7 0
t 0 20 64 71 2 5 5 89 3 27 55 0
|
| ARR18_MA0948.2_JASPAR_shift0 (ARR18:MA0948.2:JASPAR) |
 |
0.818 |
0.477 |
3.426 |
0.892 |
0.918 |
2 |
5 |
5 |
3 |
2 |
3.400 |
3 |
; cluster_5 versus ARR18_MA0948.2_JASPAR (ARR18:MA0948.2:JASPAR); m=3/6; ncol2=7; w=0; offset=0; strand=D; shift=0; score= 3.4; AGATAcG-----
; cor=0.818; Ncor=0.477; logoDP=3.426; NsEucl=0.892; NSW=0.918; rcor=2; rNcor=5; rlogoDP=5; rNsEucl=3; rNSW=2; rank_mean=3.400; match_rank=3
a 834 0 999 0 728 48 159 0 0 0 0 0
c 25 88 0 0 25 673 72 0 0 0 0 0
g 25 912 0 0 25 48 697 0 0 0 0 0
t 115 0 0 999 222 231 72 0 0 0 0 0
|
| ZFHD10-3_EEAD0065_EEADannot_shift1 (ZFHD10-3:EEAD0065:EEADannot) |
 |
0.710 |
0.592 |
5.719 |
0.878 |
0.851 |
5 |
2 |
2 |
6 |
6 |
4.200 |
4 |
; cluster_5 versus ZFHD10-3_EEAD0065_EEADannot (ZFHD10-3:EEAD0065:EEADannot); m=4/6; ncol2=10; w=0; offset=1; strand=D; shift=1; score= 4.2; -TATATATATA-
; cor=0.710; Ncor=0.592; logoDP=5.719; NsEucl=0.878; NSW=0.851; rcor=5; rNcor=2; rlogoDP=2; rNsEucl=6; rNSW=6; rank_mean=4.200; match_rank=4
a 0 10 90 10 90 10 90 10 90 10 90 0
c 0 0 0 0 0 0 0 0 0 0 0 0
g 0 0 0 0 0 0 0 0 0 0 0 0
t 0 90 10 90 10 90 10 90 10 90 10 0
|
| AtHB32.DAP_M0856_AthalianaCistrome_shift2 (AtHB32.DAP:M0856:AthalianaCistrome) |
 |
0.700 |
0.539 |
3.456 |
0.890 |
0.880 |
6 |
3 |
3 |
4 |
5 |
4.200 |
5 |
; cluster_5 versus AtHB32.DAP_M0856_AthalianaCistrome (AtHB32.DAP:M0856:AthalianaCistrome); m=5/6; ncol2=11; w=0; offset=2; strand=D; shift=2; score= 4.2; --wwwCGhATwW
; cor=0.700; Ncor=0.539; logoDP=3.456; NsEucl=0.890; NSW=0.880; rcor=6; rNcor=3; rlogoDP=3; rNsEucl=4; rNSW=5; rank_mean=4.200; match_rank=5
a 0 0 368 361 211 14 59 194 509 0 250 429
c 0 0 34 19 82 519 0 237 0 0 15 4
g 0 0 22 20 47 20 533 2 91 0 24 0
t 0 0 176 200 260 47 8 167 0 600 311 167
|
| ARR11_MA0946.1_JASPAR_rc_shift5 (ARR11:MA0946.1:JASPAR_rc) |
 |
0.815 |
0.439 |
2.644 |
0.879 |
0.897 |
3 |
6 |
6 |
5 |
3 |
4.600 |
6 |
; cluster_5 versus ARR11_MA0946.1_JASPAR_rc (ARR11:MA0946.1:JASPAR_rc); m=6/6; ncol2=8; w=0; offset=5; strand=R; shift=5; score= 4.6; -----CGTATCT
; cor=0.815; Ncor=0.439; logoDP=2.644; NsEucl=0.879; NSW=0.897; rcor=3; rNcor=6; rlogoDP=6; rNsEucl=5; rNSW=3; rank_mean=4.600; match_rank=6
a 0 0 0 0 0 13 162 242 987 4 11 32
c 0 0 0 0 0 968 2 4 2 4 982 29
g 0 0 0 0 0 5 833 22 3 2 2 11
t 0 0 0 0 0 14 3 732 7 989 5 927
|






