RSA-tools - Tutorials
The aim of these tutorials is to give a theoretical and practical introduction to the Regulatory Sequence Analysis Tools (RSAT) software suite. The most convenient way to follow the tutorial is to display the current page in a separate window, and to use the tools with the current one.
The RSAT home page displays two frames. The frame on the left contains a menu, presenting the available tools. Each time you click on a tool name, the right frame displays the form for the corresponding tool.
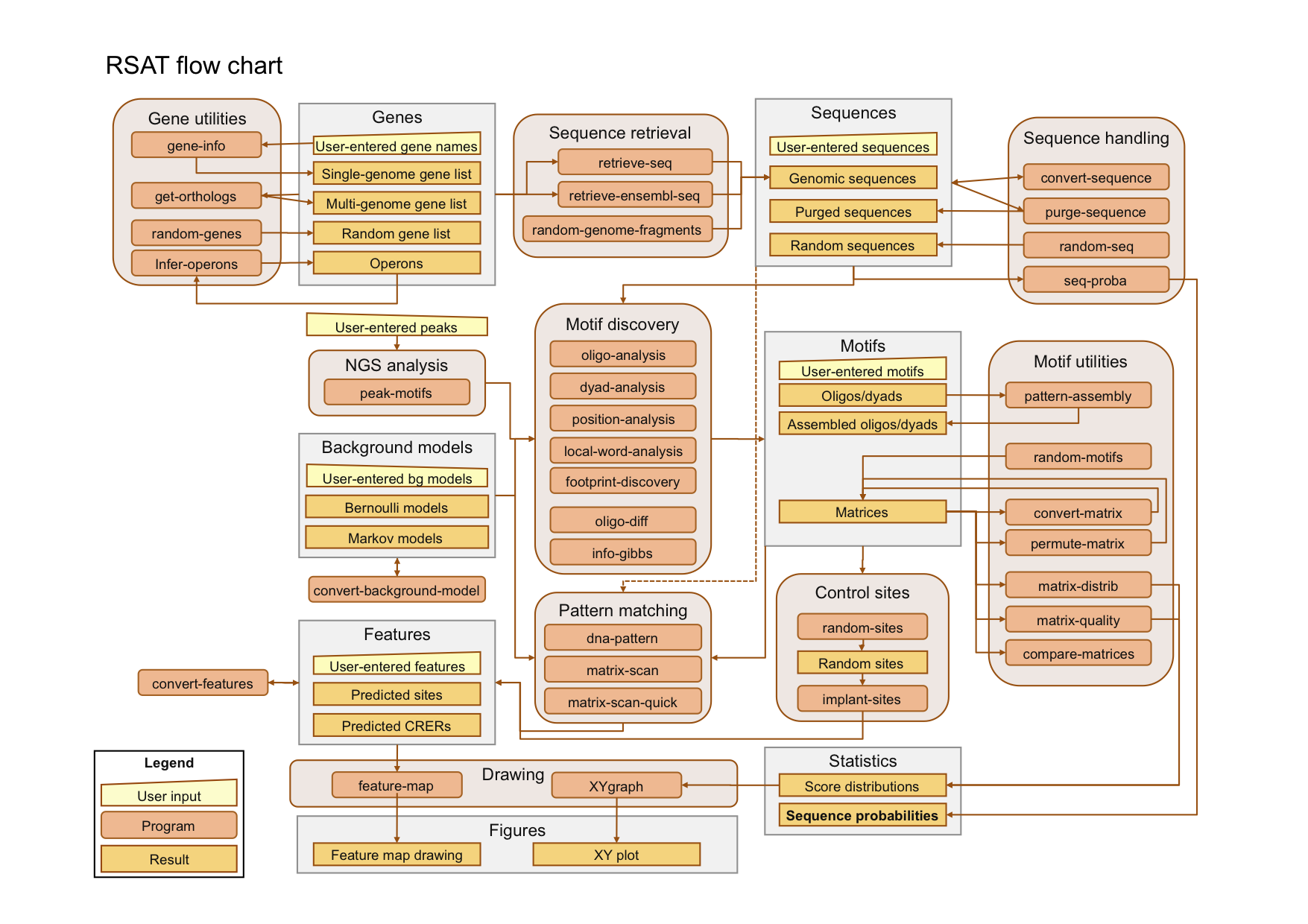
The tools are organized in a modular way : rather than having a single form for the complete analysis, we found it more convenient to present separate forms for the successive steps of a given analysis. A typical analysis will thus consist in using successivbely different tools (for example sequence retrieval -> motif discovery -> pattern matching -> feature-map). For this purpose, the tools are interconnected, allowing you to send automatically the result of one request as input for the next request (piping). The links between tools are illustrated in the flow chart below. An advantage of this modular organization is that you can either follow a full pipeline throught the tools, or directly enter at any step of an analysis with external data of your own.
We will analyze some practical examples to get familiar with the different tools, and the way they are interconnected.
The tutorial contains different parts, illustrating the typical situations that can be encountered when analysing regulatory sequences :
- Pattern matching: you know the regulatory motif (e.g. the consensus for a transcriptional factor), and you are interested by one or several particular sequences (e.g. promoters of a gene of interest, or binding fragments obtained from ChIP-on-chip experiments): you look for the matching positions within the sequences.
- Genome-scale pattern matching: you know the regulatory motif, and you would like to scan the genome to detect genes having this motif in their regulatory regions, which may be considered as potential target genes for the transcription factor of interest.
- Motif discovery (or pattern discovery). You know the sequences, you ignore the regulatory motif : you dispose of a set of functionally related regulatory sequences (e.g. promoters of co-expressed genes, or peaks collected from ChIP-seq experiments), and you suspect that they are enriched in binding site for one or seveal transcription factors. You thus want to detect a motif "ab initio" from the sequences.
Tutorials
- Abbreviation table
- How to run RSAT on the command line: Docker, Apptainer, conda
General information
- String-based representations
- Position-specific scoring matrices (PSSM)
- from RSAT
- from EnsEMBL
- dna-pattern: string-based pattern matching
- Detailed protocol for matrix-scan:
Turatsinze, J.V., Thomas-Chollier, M., Defrance, M. and van Helden, J. (2008) Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat Protoc, 3, 1578-1588. Pubmed 18802439 - Counting word occurrences in DNA sequences.
- oligo-analysis: detection of over-represented oligonucleotides (words).
- dyad-analysis: detection of over-represented spaced pairs of oligonucleotides.
- position-analysis: detection of words having a positional bias in sequences aligned on some reference position.
- Detailed protocol for string-based motif discovery:
Defrance, M., Janky, R., Sand, O. and van Helden, J. (2008) Using RSAT oligo-analysis and dyad-analysis tools to discover regulatory signals in nucleic sequences. Nature Protocols 3, 1589-1603. Pubmed 18802440 - plant upstream sequences: motif discovery on the Web browser or running a Docker container:
Ksouri N, Castro-Mondragón JA, Montardit-Tardá F, van Helden J, Contreras-Moreira B, Gogorcena Y (2021) Tuning promoter boundaries improves regulatory motif discovery in nonmodel plants: the peach example. Plant Physiol 185(3):1242-1258. Pubmed 33744946 (updates Pubmed 27557774) - matrix-clustering
- Random models
- Selecting random genes
- Generating random sequences
- Collecting peak sequences from the Galaxy Web site.
- peak-motifs: motif detection in full-size datasets of ChIP-seq peak sequences.
Representations of transcription factor binding motifs
Sequence retrieval
Pattern matching
Motif discovery
String-based motif discovery
Comparison and clustering of PSSM
Building control sets
Applications
For suggestions please post an issue on GitHub or contact the